paired end sequencing vs mate pair
Compared with inward paired-end sequencing data the outward sequencing data from mate-pair reads maybe contain the inside adapter sequence due to the random shearing. 5kb 8kb or 15kb Read.

What Is Mate Pair Sequencing For
Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.

. Ad Access more DNA discoveries than has ever before been possible with Sequencing. Mate pair reads Fragment. To simplify you can differ between two kinds of reads for paired-end sequencing.
In short-read sequencing intact genomic DNA is sheared into several million short DNA fragments called reads. The latter one is also. Paired-end sequencing facilitates detection of genomic.
Illumina Paired End Sequencing. Usually mate-pair library are used to identify structural. One of the advantages of paired end sequencing over single end is that it doubles the amount of data.
Paired-end is a type of sequencing. Illumina gets sequence data from both strands of input sequence which means it outputs data from both ends of the input and is normally reported. For classical paired-end.
In this one the molecule will be read one time from both. So if you have a 500bp fragment you machine will sequence 1-75 and 425-500 of. Get 1 month free of our Silver Membership including 2 additional DNA reports.
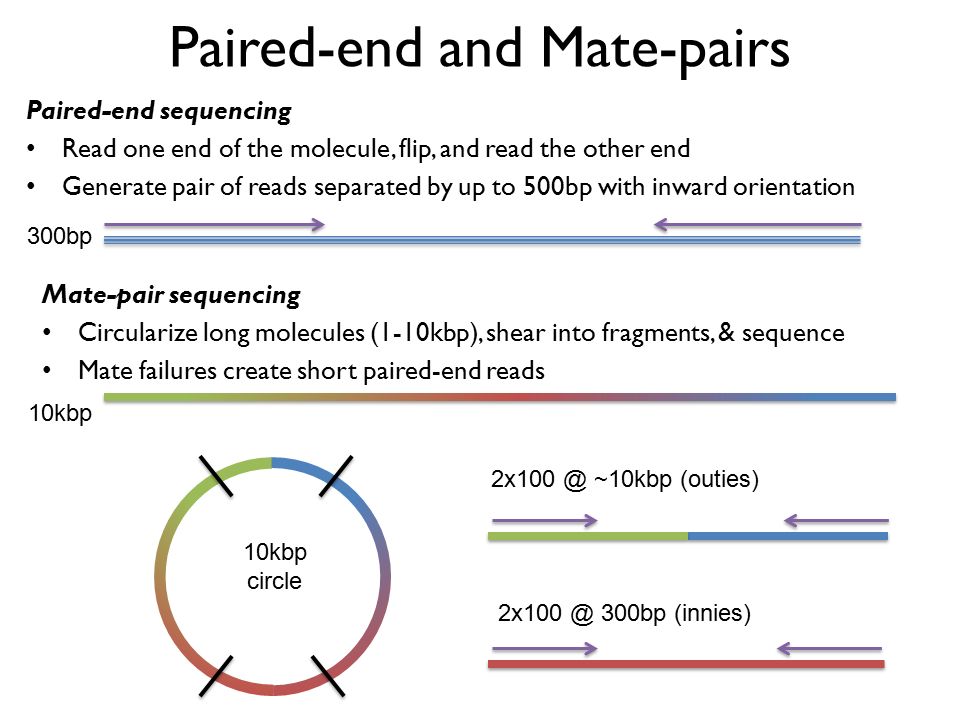
Fortunately Illumina offers paired-end PE reads which are sequences at the. In fact mate-pair libraries require paired-end sequencing. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.
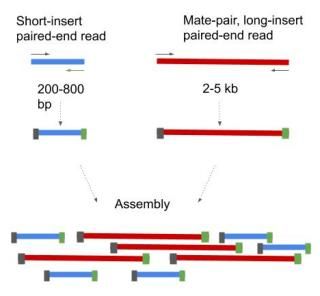
In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably. The member of a pair 1 or 2 paired-end or mate-pair reads only Versions of the Illumina pipeline since 14 appear to use NNNNNN instead of 0 for the multiplex ID. Shortinsert pairedend reads SIPERs and long-insert paired-end reads LIPERs.
What are paired end reads Illumina. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. The figure shows the.
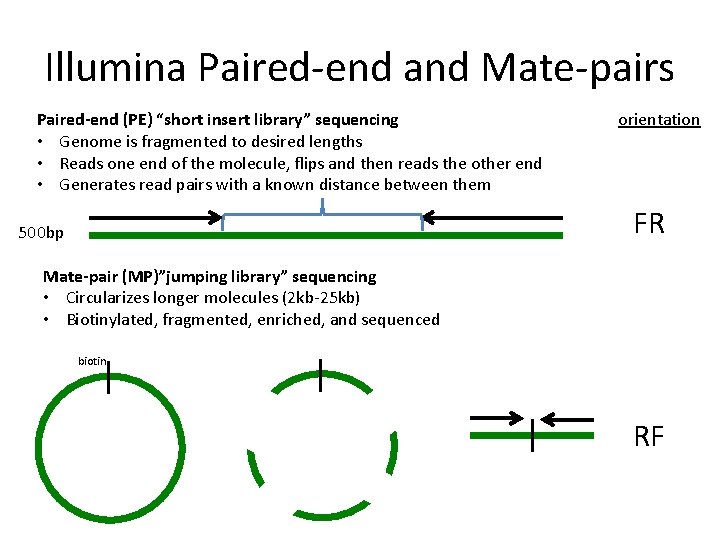
The insert size on classic paired-end is smaller about 500bp while the insert size of mate-pair is much longer several Kb which allows to join the. Paired end や mate pair という用語はどのようにライブラリが作られたかどうやってシーケンスされたかを示します. In pair-end sequencing the machine is sequencing the ends of a fragment.
While the underlying principles between PE and MP reads. Paired-end tags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they exist together only once in a genome therefore making the sequence of the. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
This can be done using either optical mapping or mate-pair sequencing. In the range of kb ie 1kb 2kb or longer in some cases. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
Also these libraries have insert sizes much longer than the paired end. Individual reads can be paired together to create paired. Paired-end sequencing facilitates detection of genomic.
Introduction to Mate Pair Sequencing. The processing of data from single-end sequencing are quite easy because there is one file per sample. Scaffolds NNN Contigs Illumina Reads Assembly Strategy Paired-end reads.
They are not two different methods. They differ in library preparation. Another supposed advantage is that it leads to more accurate reads.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Rob Edwards from San Diego State University wonders why paired-end sequencing helps with DNA sequence assembly. When you align them to the genome one read should align to the forward strand and the other should align to the reverse strand at a higher base pair position than the first one.
Paired-end sequencing facilitates detection of genomic. Mate-pair is a specific type of library.

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download Scientific Diagram

Construction Of Mate Pair Libraries Download Scientific Diagram
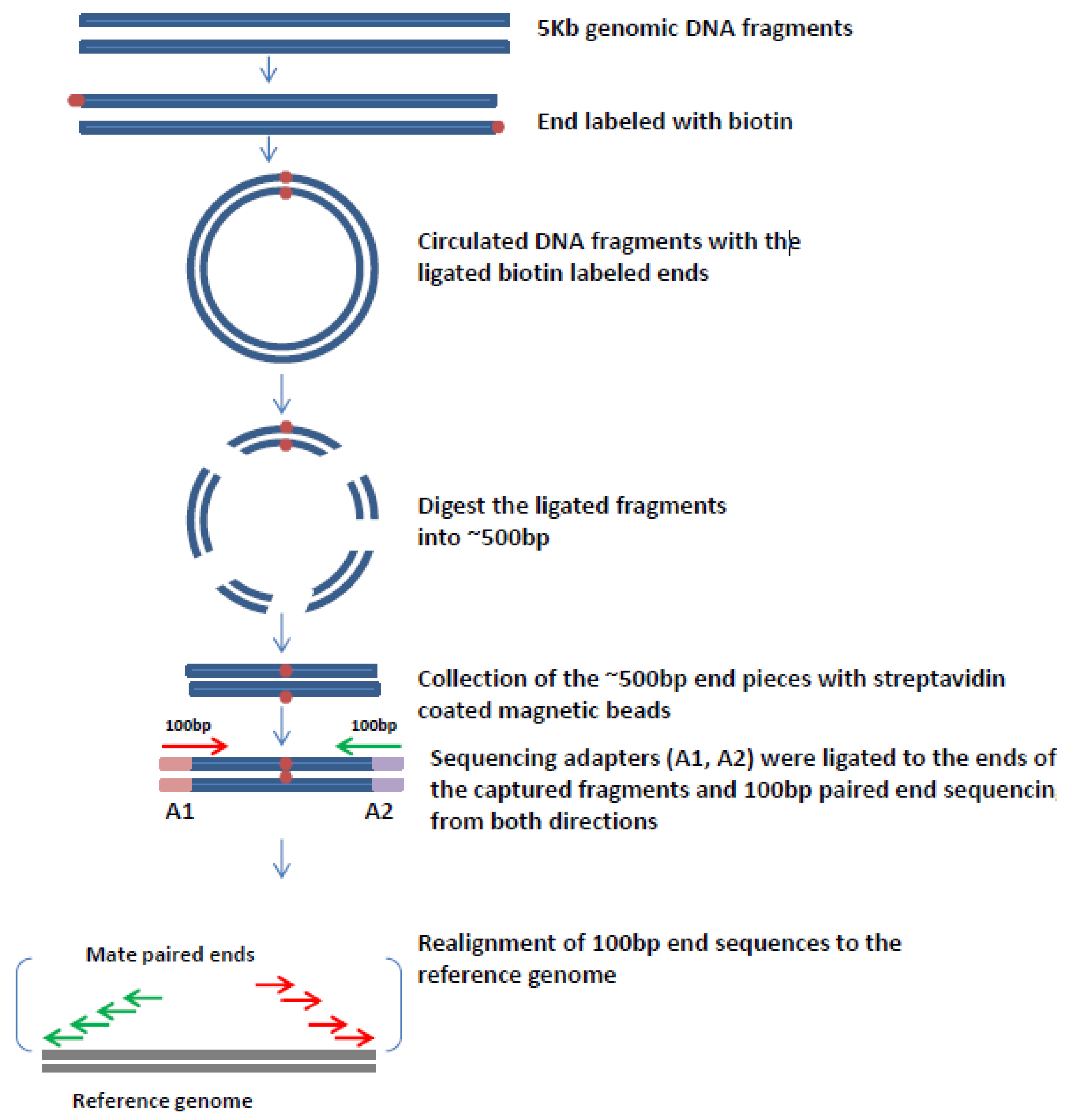
Viruses Free Full Text Mate Pair Sequencing As A Powerful Clinical Tool For The Characterization Of Cancers With A Dna Viral Etiology Html
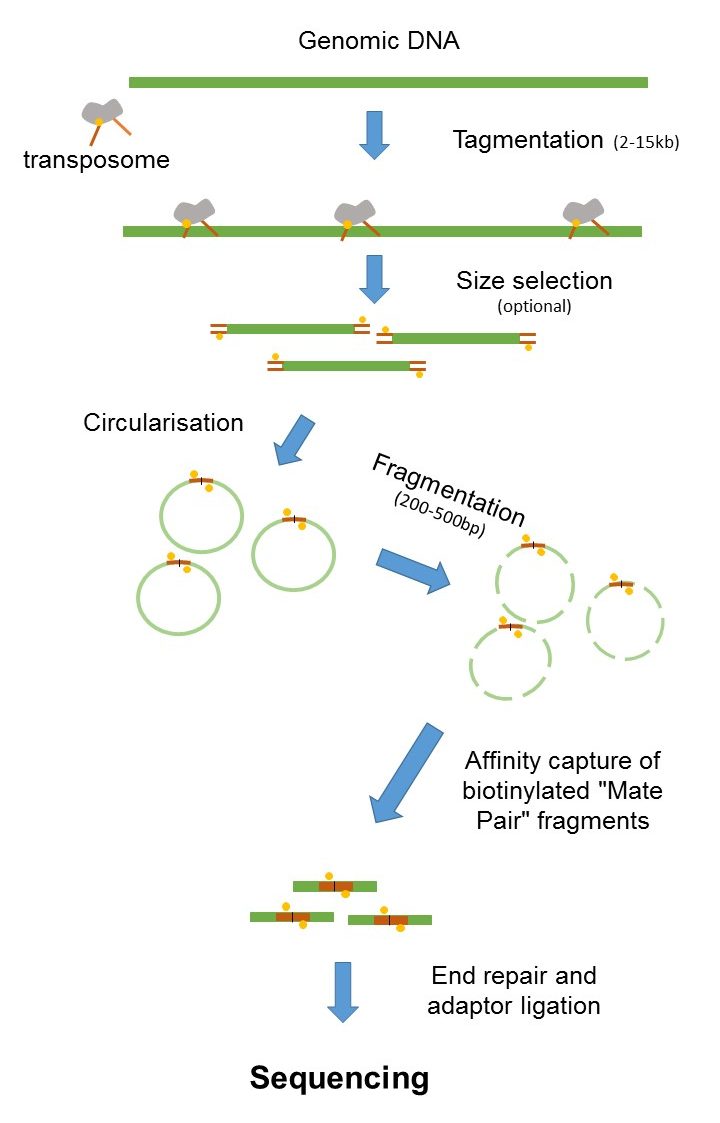
Mate Pair Sequencing France Genomique
De Novo Genome Assembly Using Next Generation Sequence

Why Do Paired Ends Map To Same Coordinates Of A Scaffold
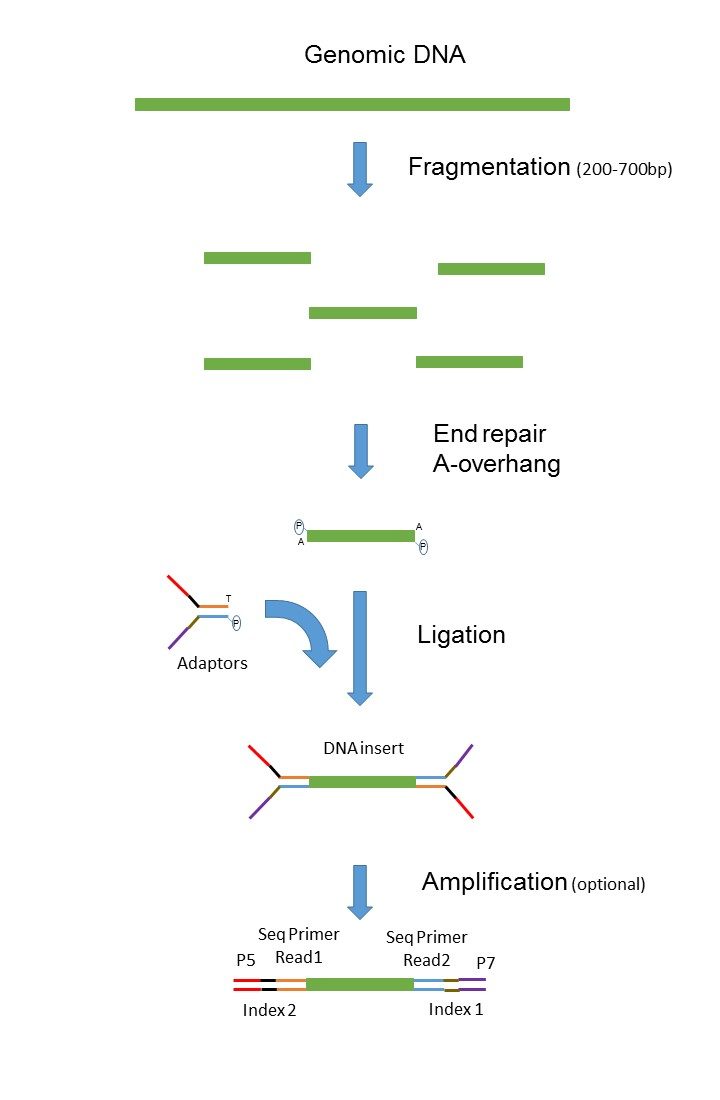
Paired End Sequencing France Genomique

6 Pairwise Strand Of Paired End And Mate Pair Read Libraries Download Scientific Diagram

Paired End Vs Single End Sequencing Reads Youtube
Whole Genome Assembly With Iplant Ppt Video Online Download

Principles For Construction Of Mate Pair Sequencing Libraries A Download Scientific Diagram

Pdf Data Processing Of Nextera Mate Pair Reads On Illumina Sequencing Platforms Semantic Scholar
Introduction To Genome Assembly Tutorial

Schematic Representation Of Mate Pair Library Construction By Coupling Download Scientific Diagram

What Is Mate Pair Sequencing For

Mate Pair Made Possible Enseqlopedia

Ion Torrent Mate Paired Library Preparation Protocol And Sequencing Download Scientific Diagram